Research
A few highlighted projects
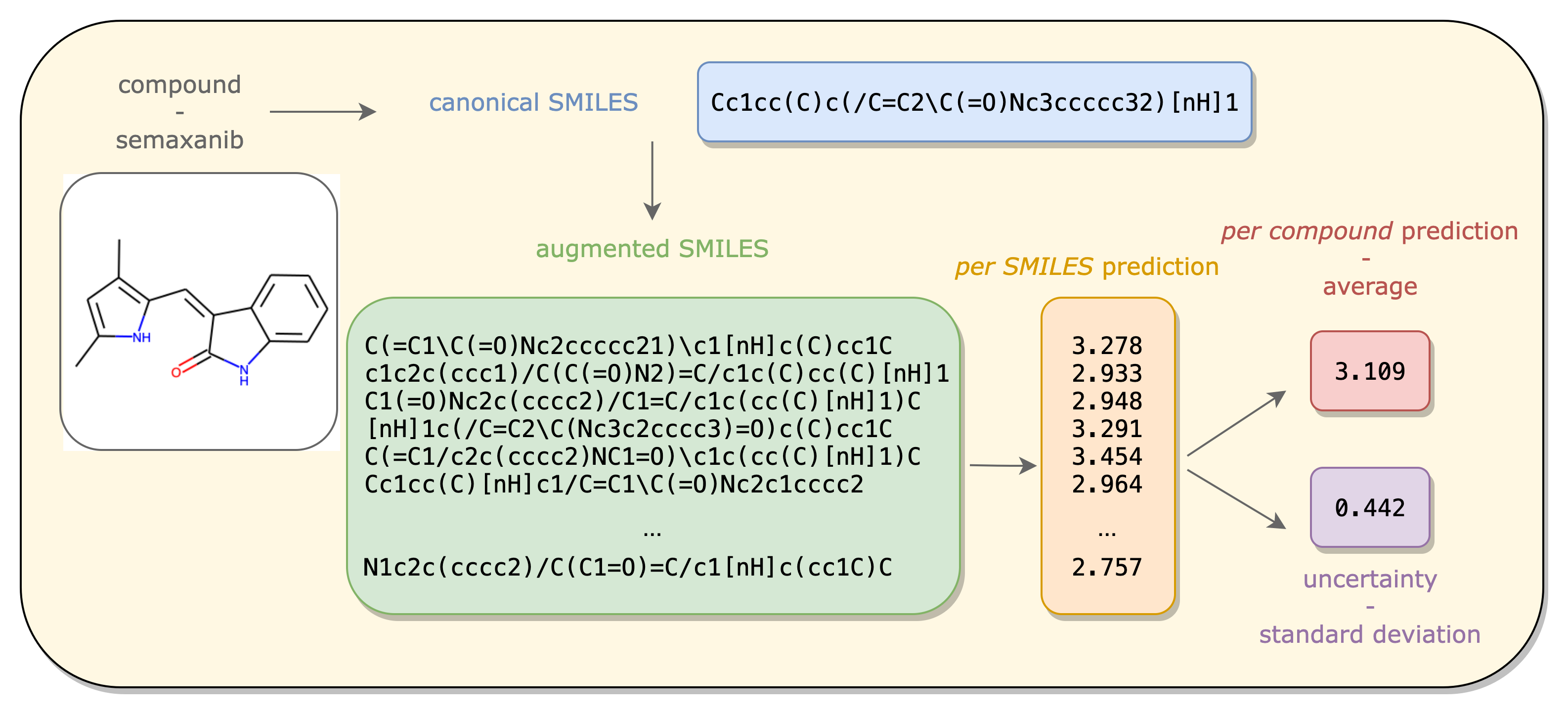
Data augmentation in deep learning
In this project, we wanted to determine if, and how would be the best way to use augmentation techniques to improve predictions in the context of drug design. We used well-known benchmark data sets to test several neural network architectures with our newly developed augmentation techniques and we beat state-of-the-art results. We also formalize the idea of confidence prediction on unlabeled data. More details can be found in the publication:
🔗: https://doi.org/10.1016/j.ailsci.2021.100014
deep learning
molecular property prediction
data augmentation
open-source
confidence assessment
SMILES
Explainable AI
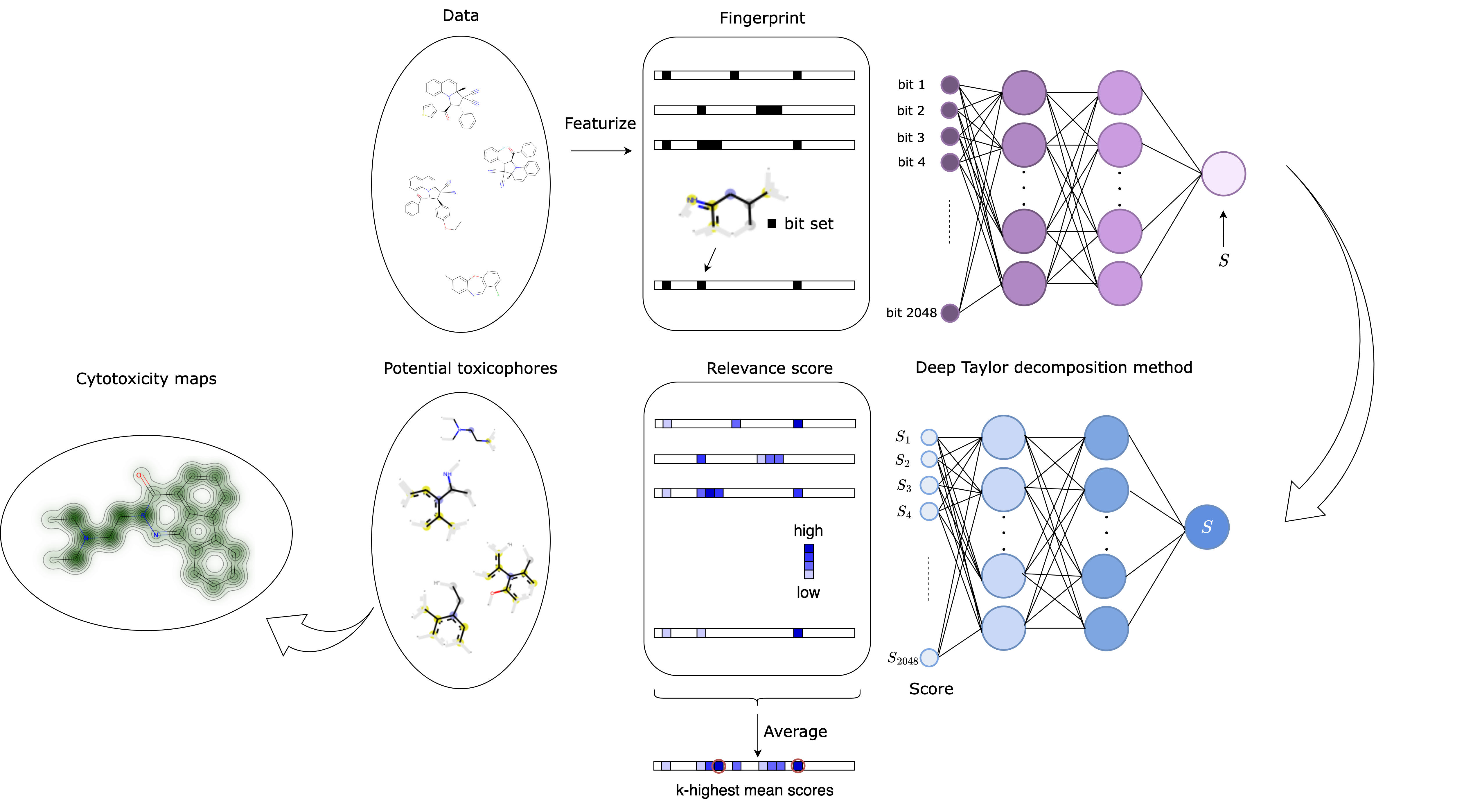
Identifying potential toxic substructures in a compound was one of the goals in this research. We used the Deep Taylor Decomposition to assign scores to atomic environments that were important in the labeling of a cytotoxic molecule. The data set came from the Leibniz Associations Research Institute for Molecular Pharmacology (FMP: Leibniz-Forschungsinstitut für Molekulare Pharmakologie), which made it highly consistent. Moreover, the percentage of compounds that were experimentally tested cytotoxic was low (approx. 4.5%), making the data set imbalanced.
deep learning
cytotoxicity maps
model interpretability
toxicity prediction
Kinase similarity assessment for off-target prediction
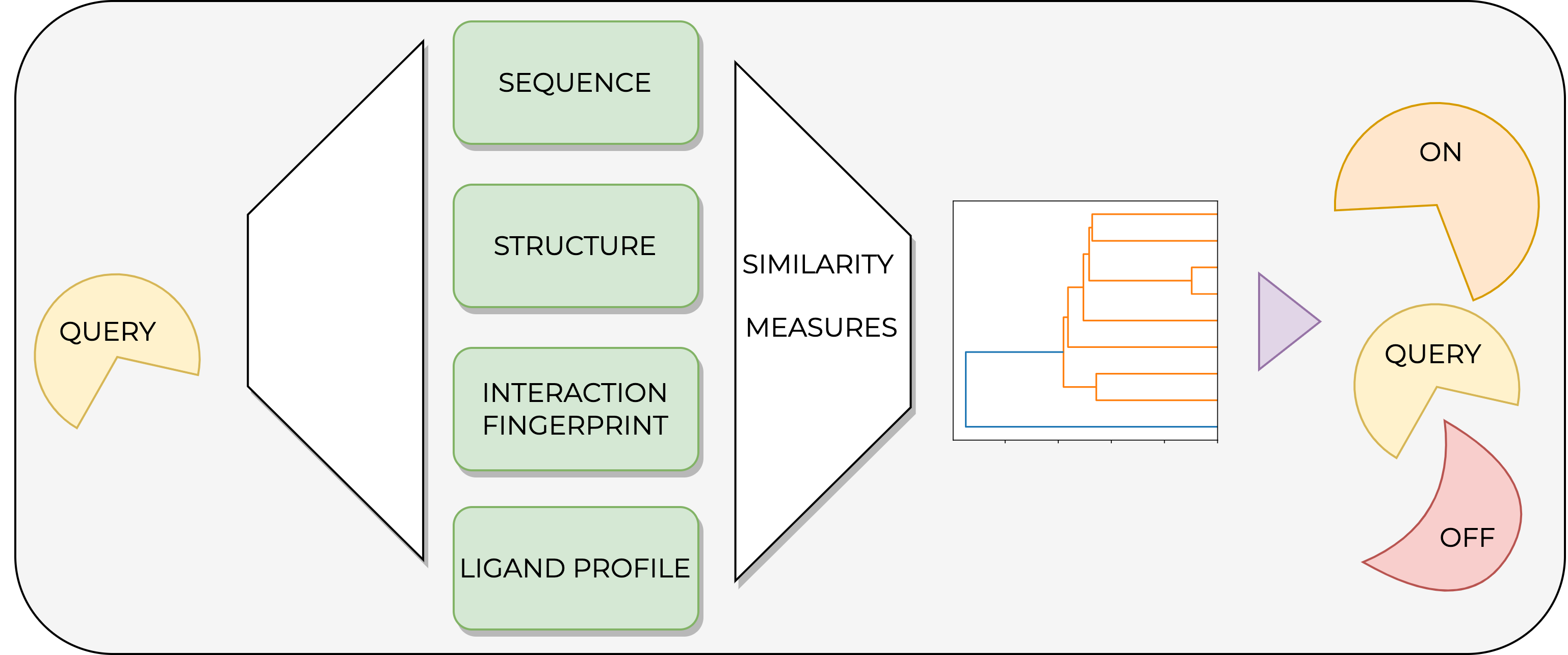
This project involves a modular pipeline that allows the comparison of kinases using four different methods, namely the pocket sequence, structural information, protein-ligand interactions, and ligand-profiling data. The proposed pipeline consists of six Jupyter notebooks. Given a set of kinases in a CSV format, four similarity measures are implemented, and kinases are compared using heatmaps and dendrograms. The project is part of TeachOpenCADD and uses open-source tools and databases such as KLIFS and ChEMBL.
kinase similarity
automated pipeline
open source
off-target prediction
Kinodata - Kinase data sets for machine learning
kinase dataset
open source
KinoML - machine learning for kinase drug discovery
kinase drug discovery
machine learnign
open source